TECHNOLOGY
miRNA inhibitors “Tough Decoy (TuD)” and “Synthetic Tough Decoy (S-TuD)”
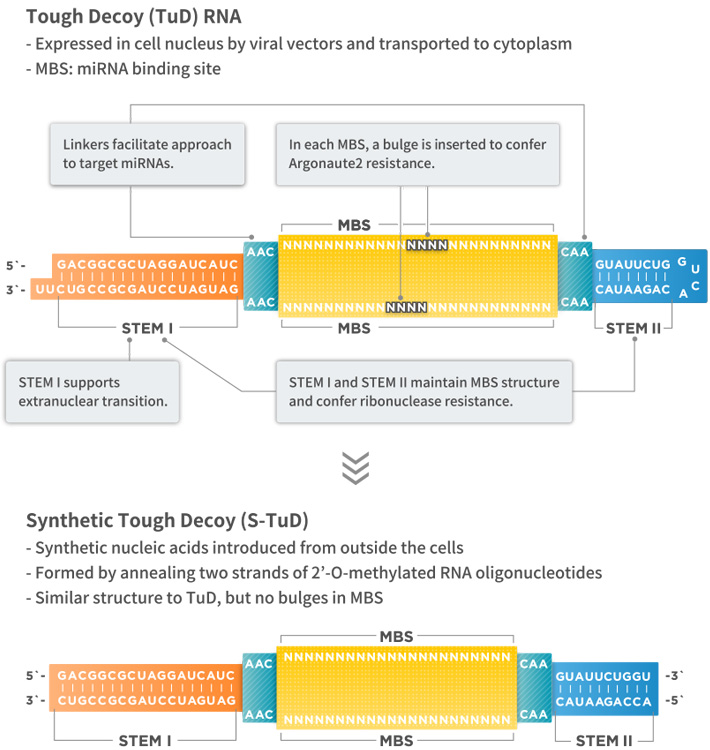
Tough Decoy (TuD) RNA expression vector is a miRNA inhibition vector that specifically inhibits the activity of target miRNAs. TuD RNAs have a unique secondary structure and consist of four functional elements: (1) STEM I of 18 bp in length; (2) two miRNA-binding sites with a sequence complementary to that of target miRNAs; (3) STEM II, a stem-loop structure connecting miRNA-binding sites; and (4) linkers connecting miRNA-binding sites and STEMs (Figure, top).
Synthetic TuD (S-TuD) is a synthetic miRNA inhibitor with a structure similar to that of the corresponding TuD RNA. It is formed by annealing two strands of 2′-O-methylated RNA oligonucleotides (Figure, bottom). TuD RNAs can be used for gene therapy, while S-TuD can apply to nucleic acid drugs.
Shematic diagram of TuD and Super-S-TuD
3 outstanding features of TuD/S-TuD:
1. Novel mechanism of action
Multiple genetic factors are intricately involved in the development and progression of most diseases. While conventional medicines target a single gene or protein, TuD/S-TuD can target multiple related genes under the control of specific miRNA.
2. Strong miRNA inhibition activity
It has been reported that the inhibitory activity of TuD/S-TuD is higher than that of conventional miRNA inhibitors*. Furthermore, TuD/S-TuD is highly resistant to ribonuclease, an RNA-degrading enzyme, and thus more stable in the blood.
* RNA. 2013 Feb;19(2):280-93.
3. Less off-target toxicity
In general, antisense oligonucleotides and small interfering RNAs (siRNAs) are prone to bind to non-target messenger RNAs with complementary sequences, causing unintended adverse effects. In contrast, TuD/S-TuD, due to topological restrictions, does not bind to non-target messenger RNAs with complementary sequences, causing few off-target effects, and the resulting toxicity is presumed to be low.
TuD/S-TuD can also be a tool to elucidate disease-specific gene regulatory networks and identify appropriate target miRNAs. miRaX has a library of TuD viral vectors for essential miRNAs. By using this library, the identification of target miRNAs can be expedited. miRaX’s scientists have long experience and insight into the gene regulatory networks formed by miRNAs; their ability to decipher the gene networks regulated by miRNAs is one of our strengths.
Publications
Haraguchi T, Nakano H, et al. A potent 2’-O-methylated RNA-based microRNA inhibitor with unique secondary structures. Nucleic Acids Res, 40; e58 (2012)
Haraguchi T, Ozaki Y, and Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res, 37: e43 (2009)


